Our broad research interests are in understanding the biochemical mechanisms that control the cell cycle. Cell growth and division are carefully coordinated by a shifting network of biomolecular interactions. Protein interactions regulate enzymatic activities responsible for key cell cycle events such as DNA replication, chromosome segregation, and cytokinesis. These events have strict spatial and temporal requirements for proper cell cycle function, and deregulation of protein interaction networks is commonly associated with aberrant cell proliferation and cancer. Understanding mechanisms of cell cycle control requires a detailed molecular picture of protein-protein interactions and how these interactions regulate enzymatic function and cellular architecture. Our laboratory seeks to elucidate the biochemical determinants of protein interaction affinity and specificity and how these factors are affected by regulatory modifications to protein composition and structure. We apply a variety of structural and biochemical techniques to learn in molecular detail how structural changes and chemical modifications affect biological function.
 Retinoblastoma Tumor Suppressor Pathway
Retinoblastoma Tumor Suppressor Pathway
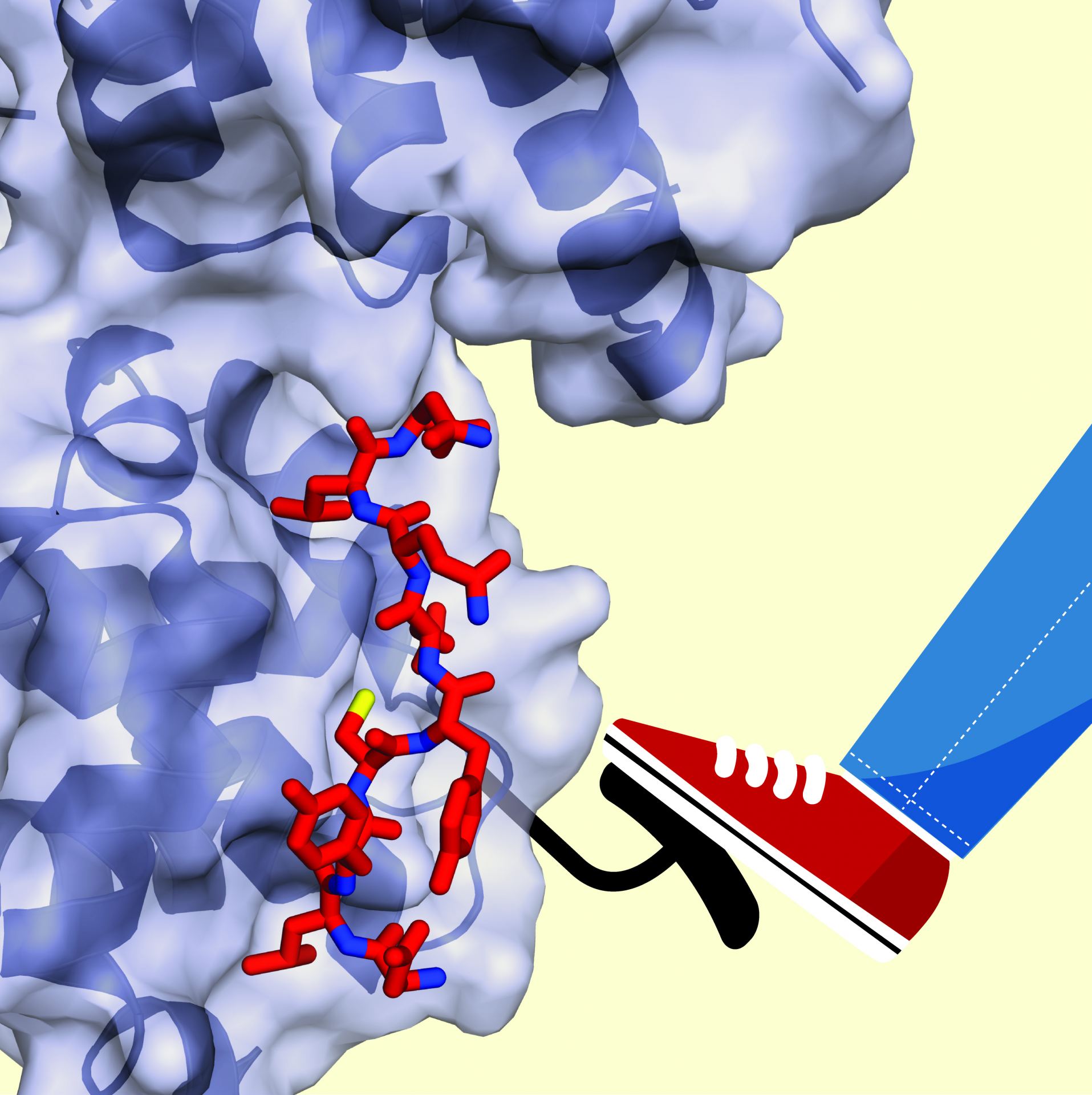
The retinoblastoma protein (Rb), which acts as a break on the cell cycle, binds and inhibits E2F transcription factors until the cell is ready to begin division. At this point, Rb is phosphorylated by cyclin-dependent kinases (Cdks) at multiple sites, E2F is released, and genes required for DNA synthesis and cell division are transcribed. The primary goal of our studies is to use atomic resolution structures and biophysical techniques to characterize the molecular interactions that stabilize the Rb-E2F complex and to understand how phosphorylation changes these interactions such that the complex dissociates. We have found that phosphorylation of different Cdk sites induces diverse structural changes in Rb to modulate its different protein interactions. We are now using our findings to develop small molecules that modulate Rb and E2F activity.
 Structure and Function of the DREAM complex
Structure and Function of the DREAM complex
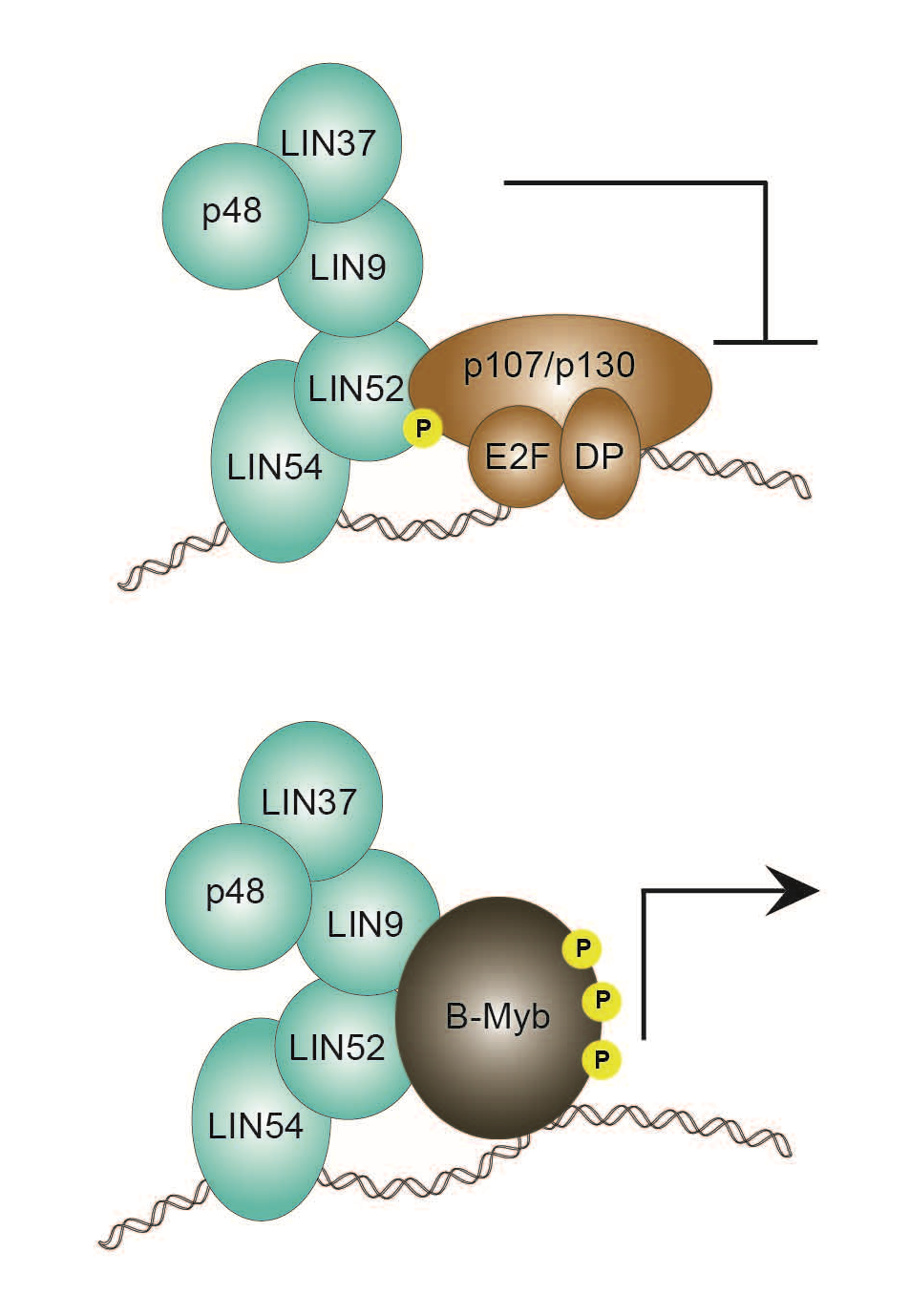
The DREAM complex is a master regulator of cell-cycle dependent gene expression. We are investigating DREAM structure to understand its biochemical function and regulation. We are characterizing how the subunits of DREAM assemble, interact with DNA, bind nucleosomes, and recruit the Myb oncoprotein to drive cell cycle progression.
 Regulation of Cyclin-Dependent Kinases
Regulation of Cyclin-Dependent Kinases
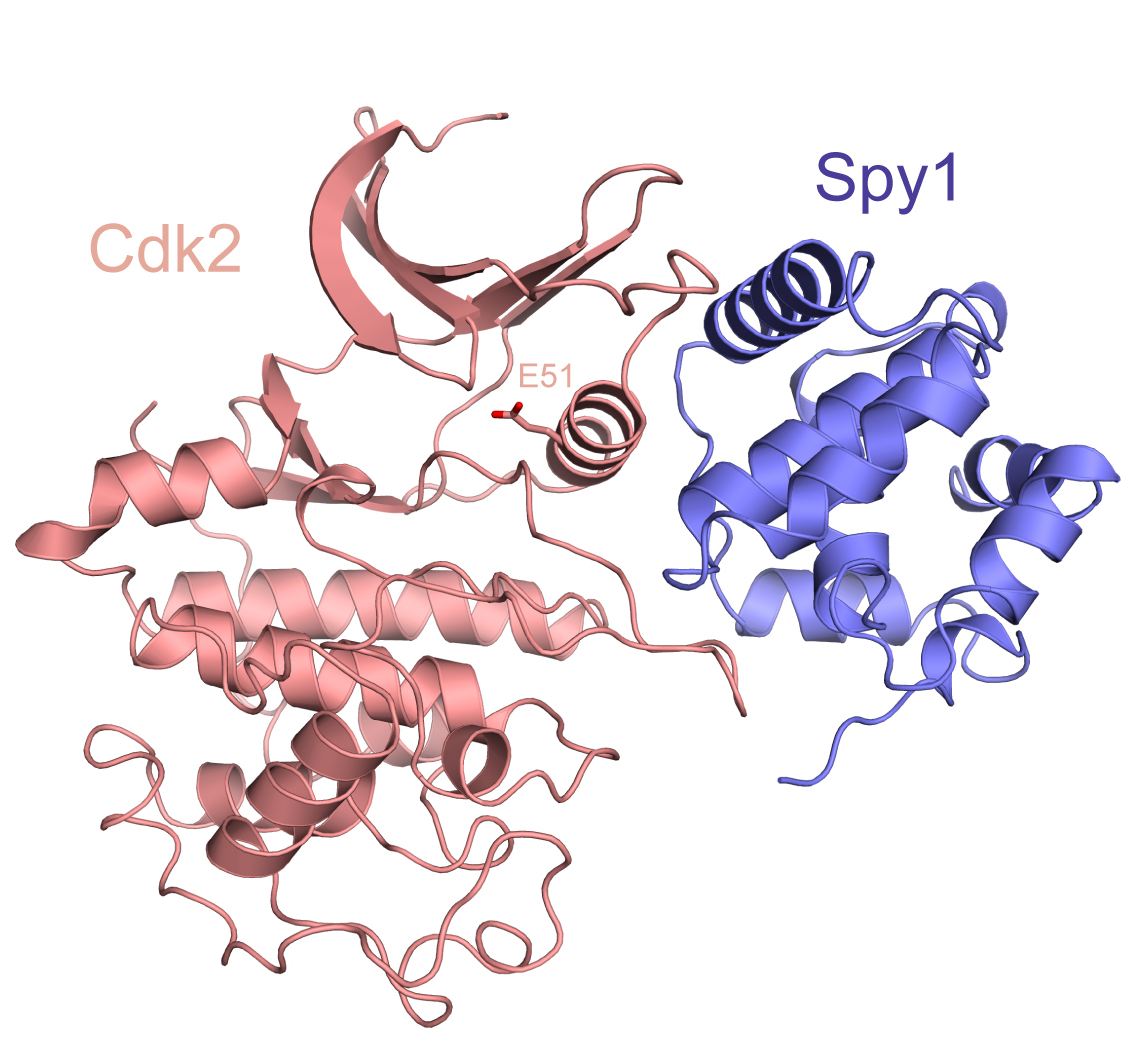
Cyclin-dependent kinases (Cdks) are the principal cell cycle regulatory enzymes that control cell fate decisions through chemical modification of a large number of protein substrates. Cdks stimulate cell cycle progression in normal and cancer cells, and so they are a significant therapeutic target for halting cell proliferation. Cdk regulation is multilayered, reflecting the need to integrate diverse growth signals to control the cell cycle. We are studying the structure and function of a number of noncanonical protein Cdk activators including Cks1, Spy1, and p21/p27. Our structural, biochemical, and cellular studies are informing how these proteins stimulate kinase function in normal and cancer cell division.
